11 Eukaryotic Cells
In this section
Content in this section includes:
Eukaryotic organisms include protozoans, algae, fungi, plants, and animals. Some eukaryotic cells are independent, single-celled microorganisms, whereas others are part of multicellular organisms. The cells of eukaryotic organisms have several distinguishing characteristics. Above all, eukaryotic cells are defined by the presence of a nucleus surrounded by a complex nuclear membrane. Also, eukaryotic cells are characterised by the presence of membrane bound organelles in the cytoplasm. Organelles such as mitochondria, the endoplasmic reticulum (ER), Golgi apparatus, lysosomes, and peroxisomes are held in place by the cytoskeleton, an internal network that supports transport of intracellular components and helps maintain cell shape (Figure 3.42). The genome of eukaryotic cells is packaged in multiple, rod-shaped chromosomes as opposed to prokaryotic cells (eg bacteria).
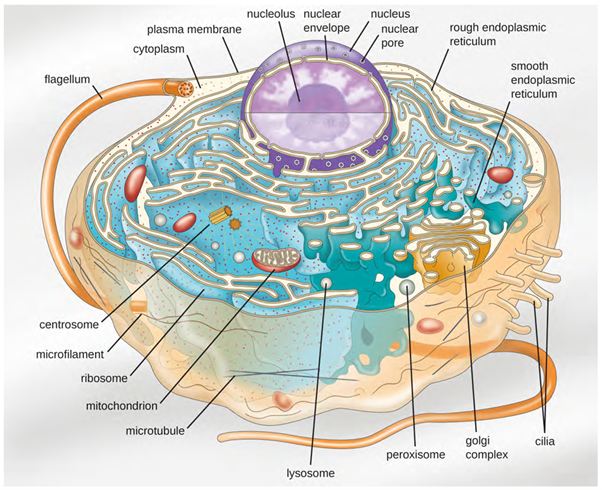
Cell Structure
Eukaryotic cells display a wide variety of different cell morphologies and shapes. Possible shapes include spheroid, ovoid, cuboidal, cylindrical, flat, lenticular, fusiform, discoidal, crescent, ring stellate, and polygonal (Figure 3.43). Some eukaryotic cells are irregular in shape, and some are capable of changing shape. The shape of a particular type of eukaryotic cell may be influenced by factors such as its primary function, the organisation of its cytoskeleton, the viscosity of its cytoplasm, the rigidity of its cell membrane or cell wall (if it has one), and the physical pressure exerted on it by the surrounding environment and/or adjoining cells. Yeast cells can change morphology and this can increase infectiveness and virulence.
Nucleus
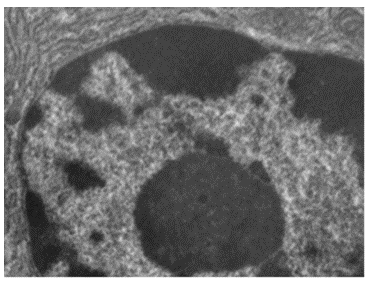
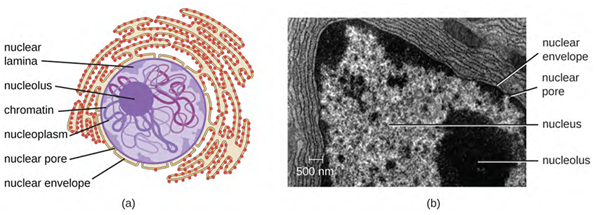
Although most eukaryotic cells have only one nucleus (Figure 3.44), exceptions exist. For example, protozoans of the genus Paramecium typically have two complete nuclei: a small nucleus that is used for reproduction (micronucleus) and a large nucleus that directs cellular metabolism (macronucleus). Additionally, some fungi transiently form cells with two nuclei, called heterokaryotic cells, during sexual reproduction. Cells whose nuclei divide, but whose cytoplasm does not, are called coenocytes.
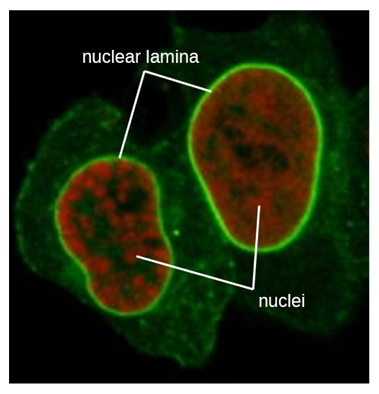
The nucleus is bound by a complex nuclear membrane, often called the nuclear envelope, that consists of two distinct lipid bilayers that are contiguous with each other (Figure 3.44). Despite these connections between the inner and outer membranes, each membrane contains unique lipids and proteins on its inner and outer surfaces. The nuclear envelope contains nuclear pores, which are large, rosette-shaped protein complexes that control the movement of materials into and out of the nucleus. The overall shape of the nucleus is determined by the nuclear lamina (Figure 3.45), a meshwork of intermediate filaments found just inside the nuclear envelope membranes. Outside the nucleus, additional intermediate filaments form a looser mesh and serve to anchor the nucleus in position within the cell.
Nucleolus
The nucleolus is a dense region within the nucleus where ribosomal RNA (rRNA) biosynthesis occurs. In addition, the nucleolus is also the site where assembly of ribosomes begins. Pre-ribosomal complexes are assembled from rRNA and proteins in the nucleolus; they are then transported out to the cytoplasm, where ribosome assembly is completed (Figure 3.46).
Endomembrane System
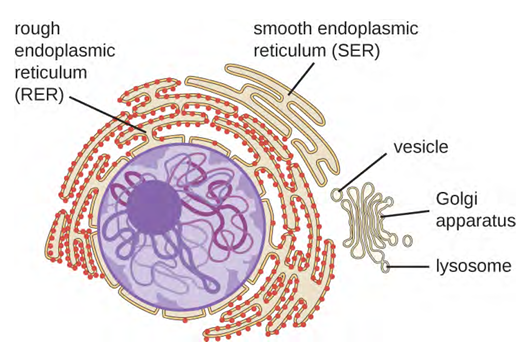
The endomembrane system, unique to eukaryotic cells, is a series of membranous tubules, sacs, and flattened discs that synthesise many cell components and move materials around within the cell (Figure 3.47). Because of their larger cell size, eukaryotic cells require this system to transport materials that cannot be dispersed by diffusion alone. The endomembrane system comprises several organelles and connections between them, including the endoplasmic reticulum, Golgi apparatus, lysosomes, and vesicles.
Endoplasmic Reticulum
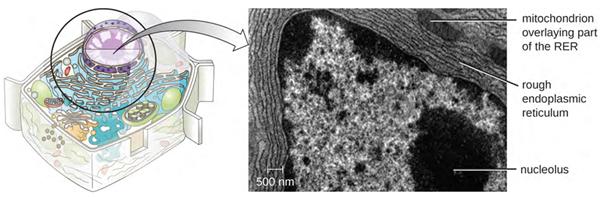
The endoplasmic reticulum (ER) is an interconnected array of tubules and cisternae (flattened sacs) with a single lipid bilayer (Figure 3.48). The spaces inside of the cisternae are called lumen of the ER. There are two types of ER, rough endoplasmic reticulum (RER) and smooth endoplasmic reticulum (SER). These two different types of ER are sites for the synthesis of distinctly different types of molecules. RER is studded with ribosomes bound on the cytoplasmic side of the membrane. These ribosomes make proteins designed for the plasma membrane (Figure 3.48). Following synthesis, these proteins are inserted into the membrane of the RER. Small sacs of the RER containing these newly synthesised proteins then bud off as transport vesicles and move either to the Golgi apparatus for further processing, directly to the plasma membrane, to the membrane of another organelle, or out of the cell. Transport vesicles are single-lipid, bilayer, membranous spheres with hollow interiors that carry molecules. SER does not have ribosomes and, therefore, appears “smooth.” It is involved in biosynthesis of lipids, carbohydrate metabolism, and detoxification of toxic compounds within the cell.
Golgi Apparatus
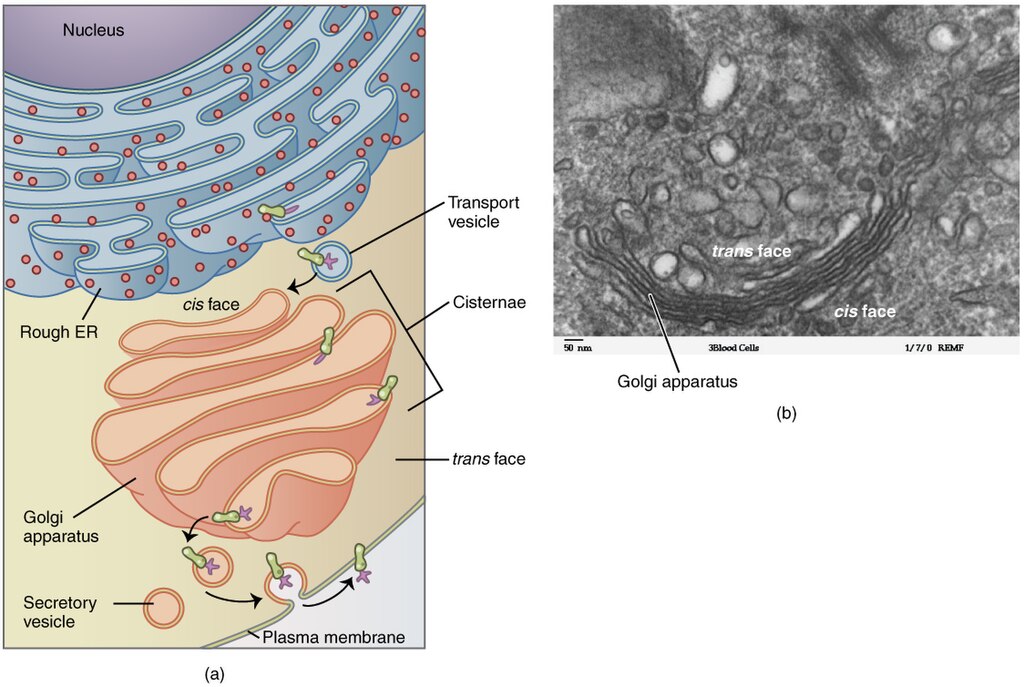
The Golgi apparatus was discovered within the endomembrane system in 1898 by Italian scientist Camillo Golgi (1843–1926), who developed a novel staining technique that showed stacked membrane structures within the cells of Plasmodium, the causative agent of malaria. The Golgi apparatus is composed of a series of membranous disks called dictyosomes, each having a single lipid bilayer, that are stacked together (Figure 3.49).
Enzymes in the Golgi apparatus modify lipids and proteins transported from the ER to the Golgi, often adding carbohydrate components to them, producing glycolipids, glycoproteins, or proteoglycans. Glycolipids and glycoproteins are often inserted into the plasma membrane and are important for signal recognition by other cells or infectious particles. Different types of cells can be distinguished from one another by the structure and arrangement of the glycolipids and glycoproteins contained in their plasma membranes. These glycolipids and glycoproteins commonly also serve as cell surface receptors.
Transport vesicles leaving the ER fuse with a Golgi apparatus on its receiving, or cis, face. The proteins are processed within the Golgi apparatus, and then additional transport vesicles containing the modified proteins and lipids pinch off from the Golgi apparatus on its outgoing, or trans, face. These outgoing vesicles move to and fuse with the plasma membrane or the membrane of other organelles.
Exocytosis is the process by which secretory vesicles (spherical membranous sacs) release their contents to the cell’s exterior (Figure 3.49). All cells have constitutive secretory pathways in which secretory vesicles transport soluble proteins that are released from the cell continually (constitutively). Certain specialised cells also have regulated secretory pathways, which are used to store soluble proteins in secretory vesicles. Regulated secretion involves substances that are only released in response to certain events or signals. For example, certain cells of the human immune system (eg mast cells) secrete histamine in response to the presence of foreign objects or pathogens in the body. Histamine is a compound that triggers various mechanisms used by the immune system to eliminate pathogens.
Chloroplast
Plant cells and algal cells contain chloroplasts, the organelles in which photosynthesis occurs (Figure 3.50). All chloroplasts have at least three membrane systems: the outer membrane, the inner membrane, and the thylakoid membrane system. Inside the outer and inner membranes is the chloroplast stroma, a gel-like fluid that makes up much of a chloroplast’s volume, and in which the thylakoid system floats. The thylakoid system is a highly dynamic collection of folded membrane sacs. It is where the green photosynthetic pigment chlorophyll is found and the light reactions of photosynthesis occur. In most plant chloroplasts, the thylakoids are arranged in stacks called grana (singular: granum), whereas in some algal chloroplasts, the thylakoids are free-floating.
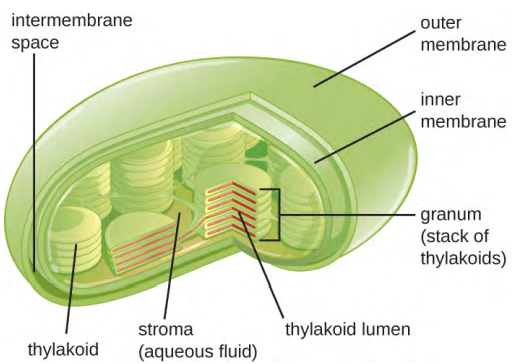
Other organelles similar to mitochondria have arisen in other types of eukaryotes, but their roles differ. Hydrogenosomes are found in some anaerobic eukaryotes and serve as the location of anaerobic hydrogen production. Hydrogenosomes typically lack their own DNA and ribosomes. Kinetoplasts are a variation of the mitochondria found in some eukaryotic pathogens. In these organisms, each cell has a single, long, branched mitochondrion in which kinetoplast DNA, organised as multiple circular pieces of DNA, is found concentrated at one pole of the cell.
Energy Production
The large, complex organelles in which aerobic cellular respiration occurs in eukaryotic cells are called mitochondria (Figure 3.51). The term “mitochondrion” was first coined by German microbiologist Carl Benda in 1898 and was later connected with the process of respiration by Otto Warburg in 1913. Scientists during the 1960s discovered that mitochondria have their own genome and 70S ribosomes. The mitochondrial genome was found to be bacterial when it was sequenced in 1976. These findings ultimately supported the endosymbiotic theory proposed by Lynn Margulis, which states that mitochondria originally arose through an endosymbiotic event in which a bacterium capable of aerobic cellular respiration was taken up by phagocytosis into a host cell and remained as a viable intracellular component.
Each mitochondrion has two lipid membranes. The outer membrane is a remnant of the original host cell’s membrane structures. The inner membrane was derived from the bacterial plasma membrane. The electron transport chain for aerobic respiration uses integral proteins embedded in the inner membrane. The mitochondrial matrix, corresponding to the location of the original bacterium’s cytoplasm, is the current location of many metabolic enzymes. It also contains mitochondrial DNA and 70S ribosomes. Invaginations of the inner membrane, called cristae, evolved to increase surface area for the location of biochemical reactions. The folding patterns of the cristae differ among various types of eukaryotic cells and are used to distinguish different eukaryotic organisms from each other.
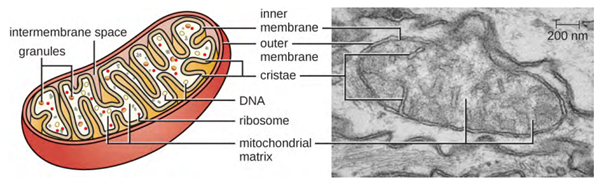
Scientists often call mitochondria (singular = mitochondrion) “powerhouses” or “energy factories” of both plant and animal cells because they are responsible for making adenosine triphosphate (ATP), the cell’s main energy-carrying molecule. ATP represents the cell’s short-term stored energy. Cellular respiration is the process of making ATP using the chemical energy in glucose and other nutrients. In mitochondria, this process uses oxygen and produces carbon dioxide as a waste product. In fact, the carbon dioxide exhaled with every breath comes from the cellular reactions and metabolic functions that produce carbon dioxide as a by-product (waste).
In keeping with the theme of form following function, it is important to point out that muscle cells have a very high concentration of mitochondria that produce ATP. Skeletal muscle cells need considerable energy to keep the body moving. When the skeletal muscle cells do not receive enough oxygen, they cannot create large amounts of ATP. Instead, production of lactic acid accompanies the small amount of ATP made by skeletal muscle cells in the absence of oxygen.
Energy Production (ATP)
Adenosine triphosphate (ATP) is the energy currency for cellular processes (Figure 3.52). ATP provides the energy for both energy-consuming endergonic reactions and energy-releasing exergonic reactions, which require a small input of activation energy. When the chemical bonds within ATP are broken, energy is released and can be harnessed for cellular work. The more bonds in a molecule, the more potential energy it contains. Because the bond in ATP is so easily broken and reformed, ATP is like a rechargeable battery that powers cellular process ranging from DNA replication to protein synthesis.
Adenosine triphosphate (ATP) is comprised of the molecule adenosine bound to three phosphate groups. Adenosine is a nucleoside consisting of the nitrogenous base adenine and the five-carbon sugar ribose. The three phosphate groups, in order of closest to furthest from the ribose sugar, are labelled alpha, beta, and gamma. Together, these chemical groups constitute an energy powerhouse. The two bonds between the phosphates are equal high-energy bonds (phosphoanhydride bonds) that, when broken, release sufficient energy to power a variety of cellular reactions and processes. The bond between the beta and gamma phosphate is considered “high-energy” because when the bond breaks, the products [adenosine diphosphate (ADP) and one inorganic phosphate group (Pi)] have a lower free energy than the reactants (ATP and a water molecule). ATP breakdown into ADP and Pi is called hydrolysis because it consumes a water molecule (hydro-, meaning “water”, and lysis, meaning “separation”).
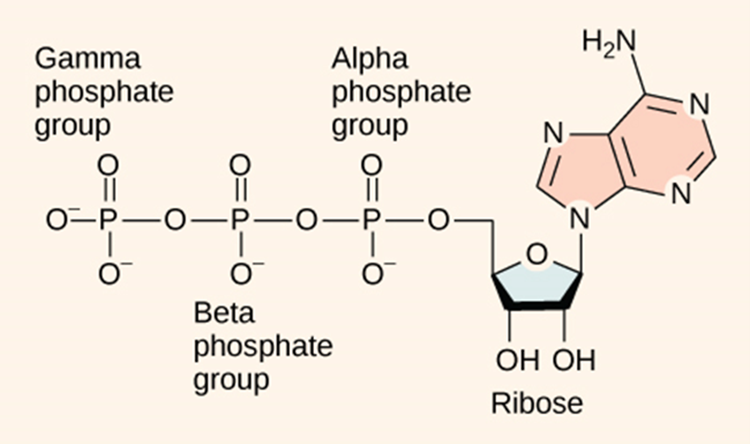
ATP is hydrolysed into ADP in the following reaction:
ATP+H2O→ADP+Pi+free energy
Like most chemical reactions, the hydrolysis of ATP to ADP is reversible. The reverse reaction combines ADP + Pi to regenerate ATP from ADP. Since ATP hydrolysis releases energy, ATP synthesis must require an input of free energy.
ADP is combined with a phosphate to form ATP in the following reaction:
ADP+Pi+free energy→ATP+H2O
Exactly how much free energy (∆G) is released with the hydrolysis of ATP, and how is that free energy used to do cellular work? The calculated ∆G for the hydrolysis of one mole of ATP into ADP and Pi is −7.3 kcal/mole (−30.5 kJ/mol). However, this is only true under standard conditions, and the ∆G for the hydrolysis of one mole of ATP in a living cell is almost double the value at standard conditions: 14 kcal/mol (−57 kJ/mol).
ATP is a highly unstable molecule. Unless quickly used to perform work, ATP spontaneously dissociates into ADP + Pi, and the free energy released during this process is lost as heat. To harness the energy within the bonds of ATP, cells use a strategy called energy coupling.
Cells couple the exergonic reaction of ATP hydrolysis with the endergonic reactions of cellular processes. For example, transmembrane ion pumps in nerve cells use the energy from ATP to pump ions across the cell membrane and generate an action potential. The sodium-potassium pump (Na+/K+pump) drives sodium out of the cell and potassium into the cell. When ATP is hydrolysed, it transfers its gamma phosphate to the pump protein in a process called phosphorylation. The Na+/K+ pump gains the free energy and undergoes a conformational change, allowing it to release three Na+ to the outside of the cell. Two extracellular K+ ions bind to the protein, causing the protein to change shape again and discharge the phosphate. By donating free energy to the Na+/K+ pump, phosphorylation drives the endergonic reaction.
During cellular metabolic reactions, or the synthesis and breakdown of nutrients, certain molecules must be altered slightly in their conformation to become substrates for the next step in the reaction series. In the very first steps of cellular respiration, glucose is broken down through the process of glycolysis. ATP is required for the phosphorylation of glucose, creating a high-energy but unstable intermediate. This phosphorylation reaction causes a conformational change that allows enzymes to convert the phosphorylated glucose molecule to the phosphorylated sugar fructose. Fructose is a necessary intermediate for glycolysis to move forward. In this example, the exergonic reaction of ATP hydrolysis is coupled with the endergonic reaction of converting glucose for use in the metabolic pathway.
Cellular Defences
Lysosomes
In the 1960s, Belgian scientist Christian de Duve (1917–2013) discovered lysosomes, membrane-bound organelles of the endomembrane system that contain digestive enzymes. Certain types of eukaryotic cells use lysosomes to break down various particles, such as food, damaged organelles or cellular debris, microorganisms, or immune complexes. Compartmentalisation of the digestive enzymes within the lysosome allows the cell to efficiently digest matter without harming the cytoplasmic components of the cell.
Peroxisomes
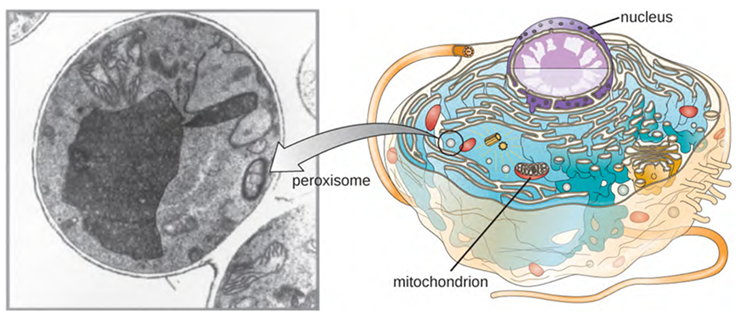
Christian de Duve is also credited with the discovery of peroxisomes, membrane-bound organelles that are not part of the endomembrane system (Figure 3.53). Peroxisomes form independently in the cytoplasm from the synthesis of peroxin proteins by free ribosomes and the incorporation of these peroxin proteins into existing peroxisomes. Growing peroxisomes then divide by a process similar to binary fission.
Peroxisomes were first named for their ability to produce hydrogen peroxide, a highly reactive molecule that helps to break down molecules such as uric acid, amino acids, and fatty acids. Peroxisomes also possess the enzyme catalase, which can degrade hydrogen peroxide. Along with the SER, peroxisomes also play a role in lipid biosynthesis. Like lysosomes, the compartmentalisation of these degradative molecules within an organelle helps protect the cytoplasmic contents from unwanted damage.
The peroxisomes of certain organisms are specialised to meet their functional needs. For example, glyoxysomes are modified peroxisomes of yeasts and plant cells that perform several metabolic functions, including the production of sugar molecules. Similarly, glycosomes are modified peroxisomes made by certain trypanosomes, the pathogenic protozoans that cause Chagas disease and African sleeping sickness.
Proteasomes
Proteasomes deal primarily with endogenous proteins; that is, proteins that were synthesised within the cell such as transcription factors, cyclins (which must be destroyed to prepare for the next step in the cell cycle) and proteins encoded by viruses and other intracellular pathogens. Proteasomes also address proteins that are folded incorrectly because of translation errors, or they are encoded by faulty genes, or they have been damaged by other molecules in the cytosol. Structure of the Proteasome in the Core Particle (CP) and the Regulatory Particle (RP) are shown in Figure 3.54.
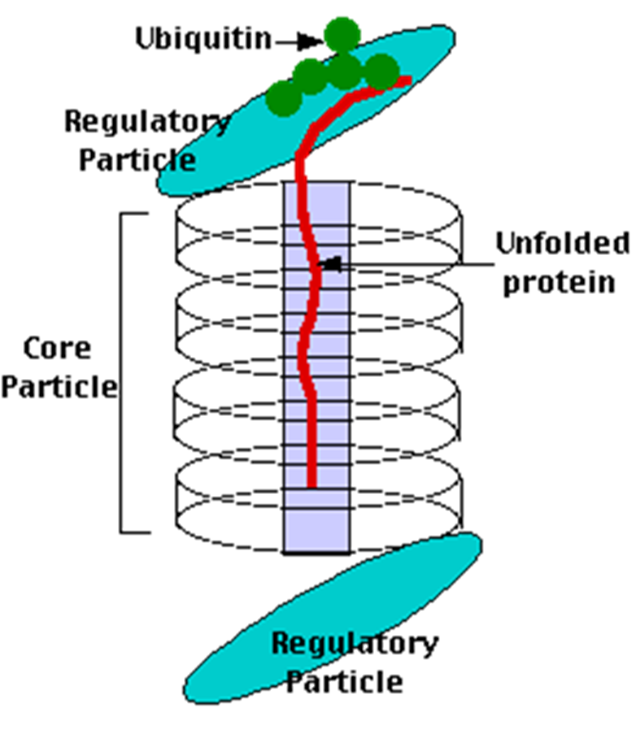
The core particle is made of 2 copies of each of 14 different proteins that are assembled in groups of 7 forming a ring. The 4 rings are stacked on each other (like 4 doughnuts) along a common centre (Figure 3.55).

There are two identical RPs, one at each end of the core particle. Each is made of 19 different proteins (none of them the same as those in the CP). 6 of these are ATPases and some of the subunits have sites that recognise the protein ubiquitin. Ubiquitin is a small protein (76 amino acids) that is conserved throughout all the kingdoms of life (Figure 3.56) and is virtually identical in sequence whether in bacteria, yeast, or mammals. Ubiquitin is used by all these creatures to target proteins for destruction (hence the name based on the word “ubiquitous”).
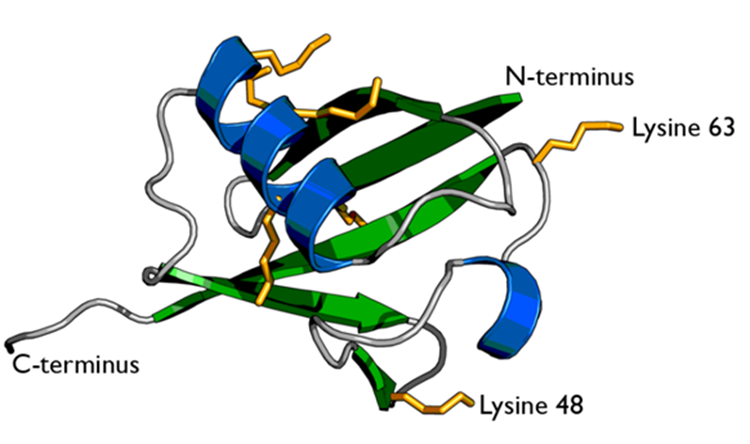
Proteins destined for destruction are conjugated to a molecule of ubiquitin which binds to the terminal amino group of a lysine residue. Additional molecules of ubiquitin bind to the first forming a chain and this complex then binds to ubiquitin-recognising site(s) on the regulatory particle. The protein is unfolded by the ATPases using the energy of ATP, which is translocated into the central cavity of the core particle. Several active sites on the inner surface of the two middle “doughnuts” break various specific peptide bonds of the chain, which produces a set of peptides averaging about 8 amino acids long. These leave the core particle by an unknown route where they may be further broken down into individual amino acids by peptidases in the cytosol. However, in mammals, they may be incorporated in a class I histocompatibility molecule to be presented to the immune system as a potential antigen. The regulatory particle releases the ubiquitins for reuse.
In mammals, activation of the immune system leads to the release of the cytokine interferon-gamma. This causes three of the subunits in the core particle to be replaced by substitute subunits; the peptides generated in this altered proteasome are picked up by TAP (= transporter associated with antigen processing) proteins and transported from the cytosol into the endoplasmic reticulum where each enters the groove at the surface of a class I histocompatibility molecule. This complex then moves through the Golgi apparatus and is inserted in the plasma membrane where it can be “recognised” by CD8+ T cells. It is probably no coincidence that the genes encoding the three substitute core particle subunits, TAP and all the MHC (major histocompatibility complex) molecules are clustered together on the same chromosome.
Copyright Information: Sources from which this module has been adapted from can be found here.
Plants, animals, protozoans, algae and fungi.
Gel-like substance composed of water and dissolved chemicals.
Large, complex organelles in which aerobic cellular respiration occurs in eukaryotic cells. Often referred to as the ‘powerhouse’ of the cell.
Interconnected array of tubules and cisternae with single lipid bilayer. Made up of rough endoplasmic reticulum (RER) and smooth endoplasmic reticulum (SER).
Stacked membranous disks called dictyosomes where lipids and proteins are modified by enzymes and repackaged for transport within the cell, to the cell membrane for use or for exocytosis from the cell.
Membrane-bound organelles of endomembrane system containing digestive enzymes.
Membrane-bound organelles that form independently in the cytoplasm from synthesis of peroxin proteins by free ribosomes and incorporation of these proteins in existing peroxisomes which then divide to multiply.
Gel-like substance composed of water and dissolved chemicals.
Full complement of DNA within a cell organised into smaller, discrete units called genes, arranged on chromosomes and plasmids.
Made of organised and packaged DNA in the form of genes and are found within the cell nucleus.
Science investigating the size, shape and structure of organisms
Plants, animals, protozoans, algae and fungi.
Membrane-bound organelle found in most eukaryotic cells, often considered the control center of the cell because it houses the cell's genetic material, DNA (deoxyribonucleic acid).
Complex nuclear membrane that surrounds the nucleus, consisting of two distinct lipid bilayers contiguous with each other.
Macromolecule that is nonpolar and insoluble in water.
Biological macromolecule comprised of one or more amino acid chains.
Large, rosette-shaped protein complexes that control the movement of materials into and out of the nucleus.
Found in eukaryotic cells and contains the DNA genome
Found in eukaryotic cells and contains the DNA genome.
Site of protein synthesis. Converts mRNA into amino acid chain that is folded into a protein and further processed in the Golgi apparatus.
Gel-like substance composed of water and dissolved chemicals.
Series of membranous tubules, sacs and flattened disks in eukaryotic cells that synthesise many cell components and move materials around within the cell.
Interconnected array of tubules and cisternae with single lipid bilayer. Made up of rough endoplasmic reticulum (RER) and smooth endoplasmic reticulum (SER).
Stacked membranous disks called dictyosomes where lipids and proteins are modified by enzymes and repackaged for transport within the cell, to the cell membrane for use or for exocytosis from the cell.
Catalyst in a biochemical reaction that is usually a complex or conjugated protein.
Macromolecule that is nonpolar and insoluble in water.
Biological macromolecule comprised of one or more amino acid chains.
Selectively permeable barrier that separates the interior of a cell from its external environment.
Secretory vesicles release the contents to the cell’s exterior.
Found within plant cells and algae and are the organelles where photosynthesis occurs.
Selectively permeable barrier that separates the interior of a cell from its external environment.
Coenzyme found in all lifeforms, that is important for metabolism as an energy molecule.
Simple sugar (monosaccharide) that is an important energy source in living organisms and is a component of many carbohydrates.
Coenzyme found in all lifeforms, that is important for metabolism as an energy molecule.
A chemical reaction where water is used to break down a compound, involving splitting a molecule into two parts by adding water.
Binary fission is a method of asexual reproduction used by many prokaryotic organisms, including bacteria and archaea.
smooth endoplasmic reticulum
Membrane-bound organelles of endomembrane system containing digestive enzymes.
Proteins associated with progression of cell cycle.
Also known as ATP synthases, they are a class of enzymes that catalyze the decomposition of ATP (adenosine triphosphate) into ADP (adenosine diphosphate) and a free phosphate ion.
Small protein that targets proteins for destruction.
A component of the proteasome, responsible for recognition of proteins tagged for degradation.
Any substance that induces an immune response by the body to that substance.
Eukaryotic, warm blooded, vertebrate animals characterised by having mammary glands.
Category of small proteins that are important in cell signalling.
White blood cell, leukocyte, attack pathogens as part of the adaptive immune system.
Gene cluster whose proteins present antigens to T cells

